Difference between revisions of "Outputs"
(→DRGEP species) |
(→DRGEP reaction) |
||
| Line 40: | Line 40: | ||
== DRGEP reaction == | == DRGEP reaction == | ||
| + | |||
| + | |||
| + | |||
| + | [[File:reac.png|300px|thumb|right|Exemple : List of the species with their interaction rate]] | ||
| + | |||
| + | During this step, the main program will display every global, backwards and forwards reactions of the scheme with the maximum of the interaction rates with the targeted species next to them. After that, new trajectories will be calculated with less and less reactions (until divergence with the reference trajectories), starting by the suppression of the less ranked ones. | ||
| + | |||
| + | Trajectories are then written in the repertory <code>/outputs</code> in .dat and new schemes in <code>/outputs/mechanisms</code> in .xml, with the name of the step, the nature of the file and the number of reactions indicated in the name. | ||
| + | |||
| + | Exemple : <code>TrajectoryDRGEPReactions54_Trajectories.dat</code> | ||
== Compute QSS Criteria == | == Compute QSS Criteria == | ||
Revision as of 16:54, 13 March 2017
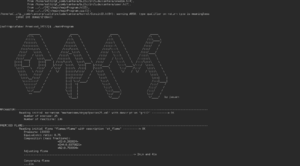
Once your "condition.cpp" is filled with the desired key words, you can run the code with
make clean
make
and
./mainProgram
If the main program is correctly running, your terminal should display the ORCh logo and all the information on your flame and your scheme.
The outputs depends on the step you are running
Contents
Compute Trajectories
This step will calculate the species trajectories without reduction.
DRGEP species
During this step, the main program will display every species of the scheme with the maximum of the interaction rates with the targeted species next to them. The interaction rates of targeted species are automatically set to 1. After that, new trajectories will be calculated with less and less species (until divergence with the reference trajectories), starting by the suppression of the less ranked ones.
Trajectories are then written in the repertory /outputs and new schemes in /outputs/mechanisms, with the name of the step, the nature of the file and the number of species indicated in the name.
Exemple : TrajectoryDRGEPSpecies29_Trajectories.dat
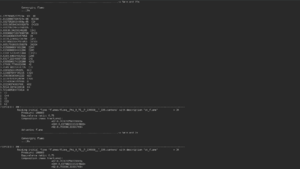
DRGEP reaction
During this step, the main program will display every global, backwards and forwards reactions of the scheme with the maximum of the interaction rates with the targeted species next to them. After that, new trajectories will be calculated with less and less reactions (until divergence with the reference trajectories), starting by the suppression of the less ranked ones.
Trajectories are then written in the repertory /outputs in .dat and new schemes in /outputs/mechanisms in .xml, with the name of the step, the nature of the file and the number of reactions indicated in the name.
Exemple : TrajectoryDRGEPReactions54_Trajectories.dat