Difference between revisions of "Multi-inlet CH 4/Air flame with scheme GRI12"
(→DRGEP Step) |
(→Optimisation Step) |
||
| Line 233: | Line 233: | ||
=== Optimisation Step === | === Optimisation Step === | ||
| + | |||
| + | |||
| + | The final step of the ORCh method, the genetic algorithm, enables to recover the trajectories of the target species. Firstly, we allowed a 10 % variation on the pre exponential factor, the temperature exposant and the energy activation in order to find their optimal values. | ||
| + | |||
| + | A population of 40 elements was used during 38 generations. | ||
| + | |||
| + | Secondly, in order to obtain more precise trajectories, we allowed a 1 % variation on the pre exponential factor, the temperature exposant and the energy activation. 42 more generations were performed in that case. | ||
| + | |||
| + | The following trajectories of the temperature, the flame speed and the target species match perfectly the ones of the reference detailed scheme with only 12 transported species and 26 reactions. | ||
| + | |||
| + | |||
| + | [[File:optim_stoch_GRI.png]] | ||
Revision as of 17:20, 25 July 2017
Objectives
The Stochastic_GRI12 test case describes a reduction of the GRI1.2 scheme for a 0D 2-inlet configuration. Starting with 32 species and 177 reactions, we reduce to 12 transported species, 2 species in quasi steady assumption and 29 reactions.
Key parameters
The target species considered for this test case are O2, CO and CO2. We are at atmospheric pressure, and the fuel is injected in its gaseous form (no evaporation model). The inputs of the 3 inlets (oxidiser, fuel and burned gases) are displayed below :
Characteristics of the air inlet :
listInlets.push_back(new MultipleInlet(
/*Temperature*/ 700,
/*Pressure*/ 1E+05,
/*Mass flow rate*/ 0.500,
/*Xk*/ "O2:0.21, N2:0.79",
/*Yk*/ "",
/*Evaporation model*/ false,
/*Droplet diameter*/ 0.0,
/*Evaporation time*/ 0.0,
/*Liquid density*/ 0.0,
/*Evaporation latent heat*/ 0.0));
Characteristics of the fuel inlet :
listInlets.push_back(new MultipleInlet(
/*Temperature*/ 450,
/*Pressure*/ 1E+05,
/*Mass flow rate*/ 0.02,
/*Xk*/ "CH4:1.0",
/*Yk*/ "",
/*Evaporation model*/ false,
/*Droplet diameter*/ 0.0,
/*Evaporation time*/ 0.0,
/*Liquid density*/ 0.0,
/*Evaporation latent heat*/ 0.0));
Characteristics of the burned gases inlet :
listInlets.push_back(new Characteristics_MultipleInlet(
/*Temperature*/ 2500,
/*Pressure*/ 1E+05,
/*Mass flow rate*/ 0.200,
/*Xk*/ "N2:0.76308, O2:0.093573, H2O:0.072355, CO2:0.070468",
/*Yk*/ "",
/*Evaporation model*/ false,
/*Droplet diameter*/ 0.0,
/*Evaporation time*/ 0.0,
/*Liquid density*/ 0.0,
/*Evaporation latent heat*/ 0.0));
/*tau_t*/ 2e-04,
/*delta_t*/ 1e-05,
/*nbIterations*/ 200,
/*BurnedGases*/ true));
Results
DRGEP Step
While running the DRGEP species step, your terminal should display the following information :
Reading initial mechanism "mechanisms/gri12.xml" with description "gri12" ----------> OK Number of species: 32 Number of reactions: 177 Set the mole fraction of inlet 0 Set the mole fraction of inlet 1 Set the mole fraction of inlet 2
Composition to enter For the equilibrium computation to get the Burned gases Compo_H_mixed 334274 X_O2: 0.16931 X_H2O: 0.019848 X_CH4: 0.0418227 X_CO2: 0.0193304 X_N2: 0.749689 T_mixed 1063.63 Nb particles 0 500 Nb particles 1 17 Nb particles 2 200 Nmix 35 Set the mole fraction of inlet 0 Set the mole fraction of inlet 1 Set the mole fraction of inlet 2
followed by the the species associated with their rank :
0 AR 2.3181e-16 C 2.44619e-09 CH 2.59722e-09 HCCOH 5.68614e-09 C2H 1.49031e-08 CH2CO 1.27776e-07 C2H2 2.68843e-07 HCCO 3.69712e-05 C2H3 7.25527e-05 CH2OH 0.00365571 H2O2 0.00537274 C2H4 0.00537274 C2H5 0.00543792 CH2(S) 0.00598963 CH2 0.0100589 CH3OH 0.0179344 H2 0.0199904 HCO 0.0576166 O 0.145908 CH3O 0.342307 H 0.373973 C2H6 0.460535 OH 0.483807 CH2O 0.999998 HO2 0.999999 CH3 1 O2 1 H2O 1 CH4 1 CO 1 CO2 1 N2
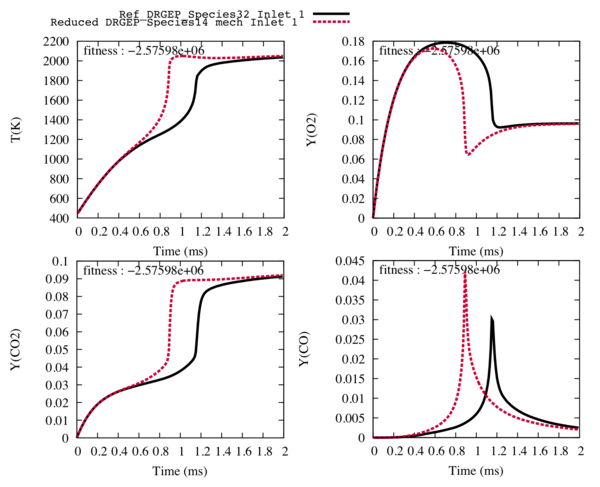
From a detailed 32 species scheme, we obtain reduced schemes (31 to 11 species). and we choose the one with 14 species for the next step. With 13 species and less, the trajectories of the targets and the temperature are too different from the detailed ones to be easily optimised.
Fig1 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 14 transported species and 42 reactions (in red).
From the reduced scheme with 14 species, the DRGEP reaction step displays the associated 42 reactions (forward, reverse and global) with their rank. The global reactions are displayed below :
Reaction 11 6.97477e-05 Reaction 2 8.75759e-05 Reaction 1 8.98678e-05 Reaction 18 0.00213229 Reaction 31 0.00291462 Reaction 6 0.00363456 Reaction 10 0.00375711 Reaction 22 0.00511265 Reaction 12 0.00898034 Reaction 7 0.0106843 Reaction 8 0.0106849 Reaction 34 0.0173608 Reaction 19 0.0185876 Reaction 25 0.0221759 Reaction 29 0.0222202 Reaction 37 0.0372901 Reaction 24 0.0446721 Reaction 9 0.0449009 Reaction 40 0.0555031 Reaction 32 0.0696855 Reaction 39 0.0865175 Reaction 3 0.144344 Reaction 30 0.156095 Reaction 14 0.166212 Reaction 35 0.167587 Reaction 38 0.178616 Reaction 23 0.250862 Reaction 36 0.357038 Reaction 21 0.360875 Reaction 26 0.419329 Reaction 28 0.459053 Reaction 5 0.487659 Reaction 42 0.602514 Reaction 20 0.633343 Reaction 15 0.740476 Reaction 16 0.789885 Reaction 17 0.828505 Reaction 41 0.846392 Reaction 33 0.948112 Reaction 13 0.969275 Reaction 4 0.971434 Reaction 27 0.992353
After this step, by comparing the reference trajectories with the new ones, we choose to delete 13 reactions, so the next step is performed with 14 species and 29 reactions. Indeed, with 28 and less species, the final state of the trajectories is not well predicted and difficult to represent even with the optimisation step.
Fig2 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 14 transported species and 29 reactions (in red).
QSS Step
While running the QSS step, your terminal should display the following information :
Species H 0.00707074 Species O 0.00347138 Species O2 1 Species OH 0.00238597 Species H2O 0.0394215 Species HO2 0.0120837 Species CH3 0.0951441 Species CH4 0.774634 Species CO 1 Species CO2 1 Species HCO 0.00480146 Species CH2O 0.178736 Species CH3O 0.00520346 Species N2 1
Interactions with species H with QSS Criteria 0.00707074 H2O:2 HO2:1 CH3:1 CH2O:2 CH3O:1
Interactions with species O with QSS Criteria 0.00347138 OH:1 H2O:1 HO2:1 CH3:1 CH2O:1 CH3O:1
Interactions with species OH with QSS Criteria 0.00238597 O:1 OH:2 HO2:1 CH3:2 HCO:2 CH2O:2 CH3O:1
Interactions with species H2O with QSS Criteria 0.0394215 H:2 O:1 HO2:1 CH3:1 HCO:2
Interactions with species HO2 with QSS Criteria 0.0120837 H:1 O:1 OH:1 H2O:1 CH3:2 CH2O:1
Interactions with species CH3 with QSS Criteria 0.0951441 H:1 O:1 OH:2 H2O:1 HO2:2 HCO:1 CH2O:1
Interactions with species HCO with QSS Criteria 0.00480146 OH:2 H2O:2 CH3:1
Interactions with species CH2O with QSS Criteria 0.178736 H:2 O:1 OH:2 HO2:1 CH3:1
Interactions with species CH3O with QSS Criteria 0.00520346 H:1 O:1 OH:1
We choose to put the species CH3O and HCO in QSS hypothesis due to their low QSS coefficient.
In order to obtain the trajectories of the 12-transported species 29-reactions reduced scheme, we run the getQSSfile step and we observe that the final state and the shape of the trajectories are conserved. It is under these conditions that the optimisation step will be efficient.
Fig3 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 12 transported species and 42 reactions (in red).
Optimisation Step
The final step of the ORCh method, the genetic algorithm, enables to recover the trajectories of the target species. Firstly, we allowed a 10 % variation on the pre exponential factor, the temperature exposant and the energy activation in order to find their optimal values.
A population of 40 elements was used during 38 generations.
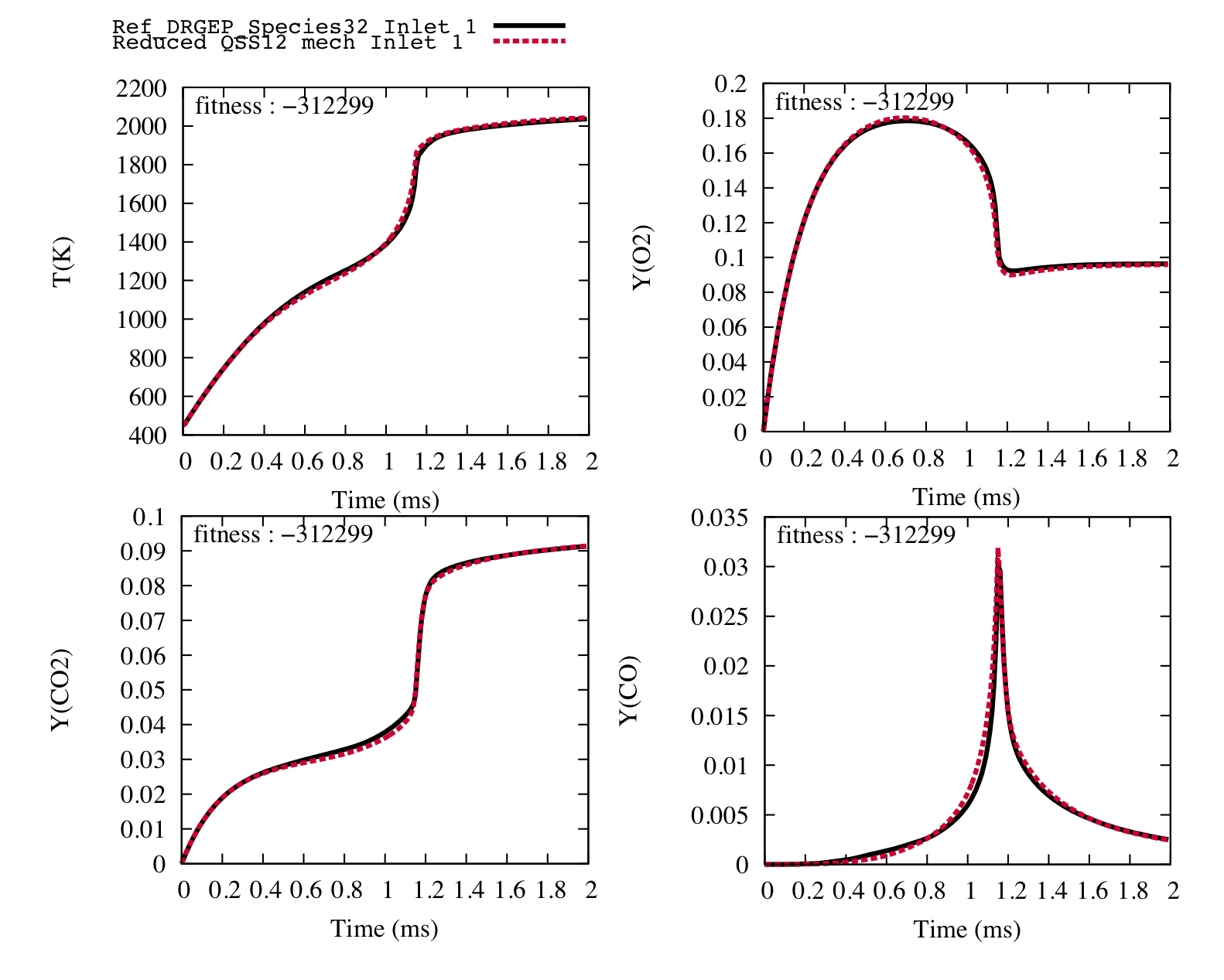
Secondly, in order to obtain more precise trajectories, we allowed a 1 % variation on the pre exponential factor, the temperature exposant and the energy activation. 42 more generations were performed in that case.
The following trajectories of the temperature, the flame speed and the target species match perfectly the ones of the reference detailed scheme with only 12 transported species and 26 reactions.