Difference between revisions of "Premix CH 4/Air flame with scheme GRI12"
(→DRGEP step) |
(→DRGEP step) |
||
| (21 intermediate revisions by 2 users not shown) | |||
| Line 6: | Line 6: | ||
| − | The target species considered for this test case are O2, CO and CO2. The characteristics of the premixed flame are displayed below : | + | The target species considered for this test case are O2, CO and CO2. The characteristics of the premixed flame are displayed below (part of the input_file.ini) : |
| − | + | //------Flame parameters------// | |
| − | + | //Flame 0 | |
| − | + | Tf = 300 | |
| − | + | To = 300 | |
| − | + | Pressure = 1E+05 | |
| − | + | Equivalence_ratio = 0.75 | |
| − | + | Xf = CH4:1.0 | |
| − | + | Xo = O2:0.21, N2:0.79 | |
| − | + | // the composition can also be added with mass fractions, for that replace "Xf" by "Yf" and "Xo" by "Yo" | |
| − | + | Initial_flame = flames/flame__Phi_0_75__P_100000__T_300.cantera | |
| + | Final_flame = flames/flame | ||
| + | //End | ||
== '''Results''' == | == '''Results''' == | ||
| Line 23: | Line 25: | ||
=== DRGEP step === | === DRGEP step === | ||
| − | While running the DRGEP species step, your terminal should display the following information : | + | While running the DRGEP species step (with <code>step = DRGEP_Species</code> in the "input_file.ini"), your terminal should display the following information : |
| + | MECHANISM:------------------------------------------------------------------------------------------------------ | ||
| + | Reading initial mechanism "mechanisms/gri12.xml" with description "gri12" ----------> OK | ||
| + | Number of species: 32 | ||
| + | Number of reactions: 177 | ||
| − | + | PREMIXED FLAME:-------------------------------------------------------------- | |
| − | + | Reading initial flame "flames/flame__Phi_0_75__P_100000__T_300.cantera" with description "st_flame" ----------> description: test flame | |
| − | + | OK | |
| − | + | Pressure: 100000 | |
| − | Pressure : | + | Equivalence ratio: 0.75 |
| − | + | Composition (mass fractions): | |
| − | + | <O2:0.223145> | |
| − | + | <CH4:0.0419532> | |
| − | + | <N2:0.734901> | |
| Line 40: | Line 46: | ||
followed by the the species associated with their rank : | followed by the the species associated with their rank : | ||
| + | -------DRGEP coefficients------- | ||
| + | -------------------------------- | ||
1.37642e-15 AR | 1.37642e-15 AR | ||
2.12795e-06 HCCOH | 2.12795e-06 HCCOH | ||
| Line 76: | Line 84: | ||
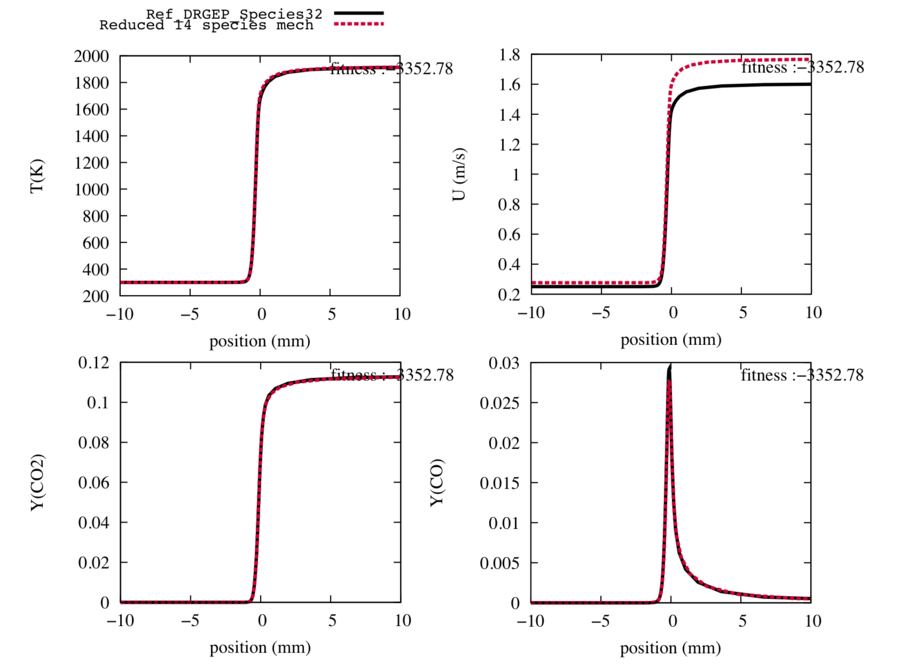
| − | From a detailed 32 species scheme, we obtain reduced schemes (31 to 14 species). and we choose the one with 14 species for the next step because the shape of the velocity and species are conserved (and Cantera is unable to converge the 13-species scheme). | + | From a detailed 32 species scheme, we obtain reduced schemes (31 to 14 species). and we choose the one with 14 species for the next step because the shape of the velocity and species are conserved (and Cantera is unable to converge the 13-species scheme. The code will therefore end with an error message. But it is fine). The following Figure 1 is available in the directory <code>outputs/Premixed/</code> with the file format of EPS. |
[[File:DRGEPSpec_14Sp_42R_Premixed.png|900px|center]] | [[File:DRGEPSpec_14Sp_42R_Premixed.png|900px|center]] | ||
| Line 82: | Line 90: | ||
Fig1 : Comparison between the reference trajectories of the target species, the temperature and the flame speed (in black), and the trajectories computed with the reduced mechanism with 14 transported species and 42 reactions, after a DRGEP species reduction (in red). | Fig1 : Comparison between the reference trajectories of the target species, the temperature and the flame speed (in black), and the trajectories computed with the reduced mechanism with 14 transported species and 42 reactions, after a DRGEP species reduction (in red). | ||
| − | |||
| − | + | From the reduced scheme with 14 species, we then perform the next DRGEP Reactions step by manually change the value of the "step" keyword in the "input_file.ini" as <code>step = DRGEP_Reactions</code>. Make sure the reference mechanism and trajectory are correctly set up (i.e., <code>mech_ref = mechanisms/gri12.xml; trajectory_ref = Ref_DRGEP_Species32</code>). The DRGEP Reactions step displays the associated 42 reactions (forward, reverse and global) with their rank : | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | |||
| + | Reaction 22 9.23227e-06 | ||
| + | Reaction 1 2.08238e-05 | ||
| + | Reaction 11 2.29985e-05 | ||
| + | Reaction 2 5.30076e-05 | ||
| + | Reaction 12 0.00029274 | ||
| + | Reaction 18 0.00191785 | ||
| + | Reaction 6 0.00198407 | ||
| + | Reaction 36 0.00293471 | ||
| + | Reaction 38 0.00420356 | ||
| + | Reaction 31 0.00462285 | ||
| + | Reaction 34 0.00611017 | ||
| + | Reaction 7 0.00905302 | ||
| + | Reaction 10 0.00921383 | ||
| + | Reaction 32 0.0106164 | ||
| + | Reaction 37 0.0133867 | ||
| + | Reaction 8 0.0136681 | ||
| + | Reaction 25 0.0237175 | ||
| + | Reaction 24 0.0277978 | ||
| + | Reaction 29 0.0305413 | ||
| + | Reaction 13 0.0329515 | ||
| + | Reaction 9 0.0342601 | ||
| + | Reaction 3 0.0398655 | ||
| + | Reaction 5 0.0422524 | ||
| + | Reaction 14 0.0442348 | ||
| + | Reaction 19 0.0493222 | ||
| + | Reaction 35 0.102885 | ||
| + | Reaction 40 0.103946 | ||
| + | Reaction 26 0.111933 | ||
| + | Reaction 30 0.131232 | ||
| + | Reaction 23 0.159996 | ||
| + | Reaction 21 0.161005 | ||
| + | Reaction 20 0.180048 | ||
| + | Reaction 39 0.18465 | ||
| + | Reaction 42 0.19675 | ||
| + | Reaction 4 0.208145 | ||
| + | Reaction 15 0.215404 | ||
| + | Reaction 16 0.222982 | ||
| + | Reaction 27 0.274358 | ||
| + | Reaction 17 0.274617 | ||
| + | Reaction 33 0.403162 | ||
| + | Reaction 41 0.437888 | ||
| + | Reaction 28 0.438952 | ||
| Line 135: | Line 144: | ||
[[File:DRGEPR_14sp_26R_Premixed.png|900px|center]] | [[File:DRGEPR_14sp_26R_Premixed.png|900px|center]] | ||
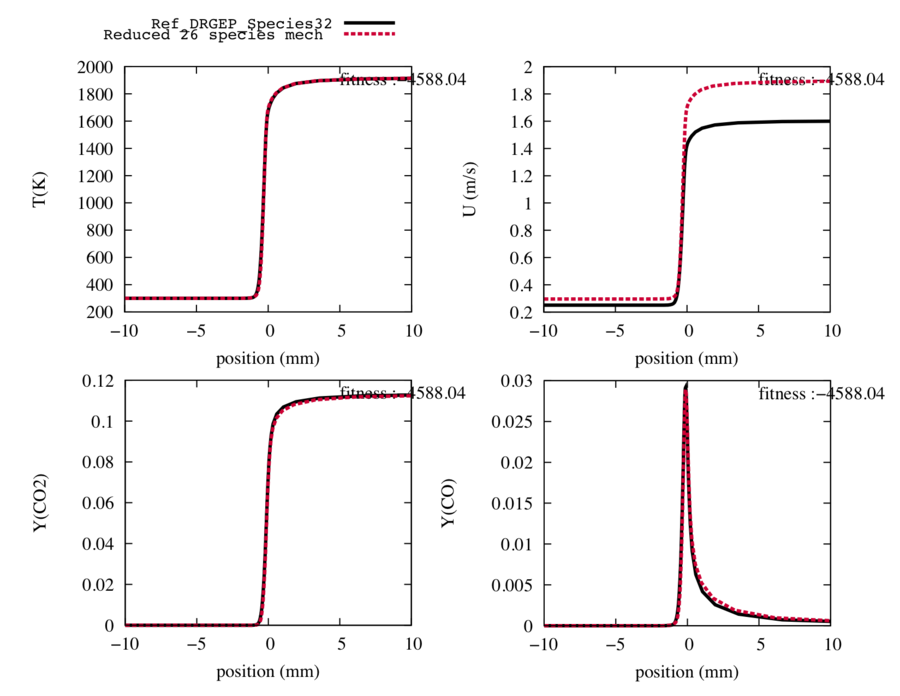
| − | Fig2 : Comparison between the trajectories of the target species, the temperature and the flame speed computed with the | + | Fig2 : Comparison between the trajectories of the target species, the temperature and the flame speed computed with the reference 32-species 177-reaction mechanism (in black) , and the trajectories computed with the reduced mechanism with 14 transported species and 26 reactions after a DRGEP reaction reduction (in red). |
=== QSS step === | === QSS step === | ||
| − | The | + | The '''computeQSSCriteria''' step provides the QSS criteria of each species and the different links with the others species : |
| − | Species H 0. | + | Species H 0.0752034 |
| − | Species O 0. | + | Species O 0.0211171 |
| − | Species O2 0. | + | Species O2 0.33194 |
| − | Species OH 0. | + | Species OH 0.019287 |
| − | Species H2O 0. | + | Species H2O 0.379235 |
| − | Species HO2 0. | + | Species HO2 0.0113201 |
| − | Species CH3 0. | + | Species CH3 0.107792 |
| − | Species CH4 0. | + | Species CH4 0.79541 |
| − | Species CO 0. | + | Species CO 0.693792 |
| − | Species CO2 0. | + | Species CO2 0.720209 |
| − | Species HCO 0. | + | Species HCO 0.00321461 |
| − | Species CH2O 0. | + | Species CH2O 0.0817911 |
| − | Species CH3O 0. | + | Species CH3O 0.0360026 |
Species N2 0 | Species N2 0 | ||
| − | Interactions with species H with QSS Criteria 0. | + | |
| + | Interactions with species H with QSS Criteria 0.0752034 | ||
HO2:2 CH3:1 CH2O:2 CH3O:1 N2:1 | HO2:2 CH3:1 CH2O:2 CH3O:1 N2:1 | ||
| − | Interactions with species O with QSS Criteria 0. | + | Interactions with species O with QSS Criteria 0.0211171 |
OH:1 HO2:1 CH3:1 CH2O:1 CH3O:1 | OH:1 HO2:1 CH3:1 CH2O:1 CH3O:1 | ||
| − | Interactions with species OH with QSS Criteria 0. | + | Interactions with species OH with QSS Criteria 0.019287 |
O:1 OH:2 HO2:1 CH3:2 HCO:2 CH2O:1 CH3O:1 | O:1 OH:2 HO2:1 CH3:2 HCO:2 CH2O:1 CH3O:1 | ||
| − | Interactions with species HO2 with QSS Criteria 0. | + | Interactions with species HO2 with QSS Criteria 0.0113201 |
H:2 O:1 OH:1 CH3:1 CH2O:1 N2:1 | H:2 O:1 OH:1 CH3:1 CH2O:1 N2:1 | ||
| − | Interactions with species CH3 with QSS Criteria 0. | + | Interactions with species CH3 with QSS Criteria 0.107792 |
H:1 O:1 OH:2 HO2:1 | H:1 O:1 OH:2 HO2:1 | ||
| − | Interactions with species HCO with QSS Criteria 0. | + | Interactions with species HCO with QSS Criteria 0.00321461 |
OH:2 | OH:2 | ||
| − | Interactions with species CH2O with QSS Criteria 0. | + | Interactions with species CH2O with QSS Criteria 0.0817911 |
H:2 O:1 OH:1 HO2:1 | H:2 O:1 OH:1 HO2:1 | ||
| − | Interactions with species CH3O with QSS Criteria 0. | + | Interactions with species CH3O with QSS Criteria 0.0360026 |
H:1 O:1 OH:1 | H:1 O:1 OH:1 | ||
Interactions with species N2 with QSS Criteria 0 | Interactions with species N2 with QSS Criteria 0 | ||
| − | H:1 HO2:1 | + | H:1 HO2:1 |
| Line 189: | Line 199: | ||
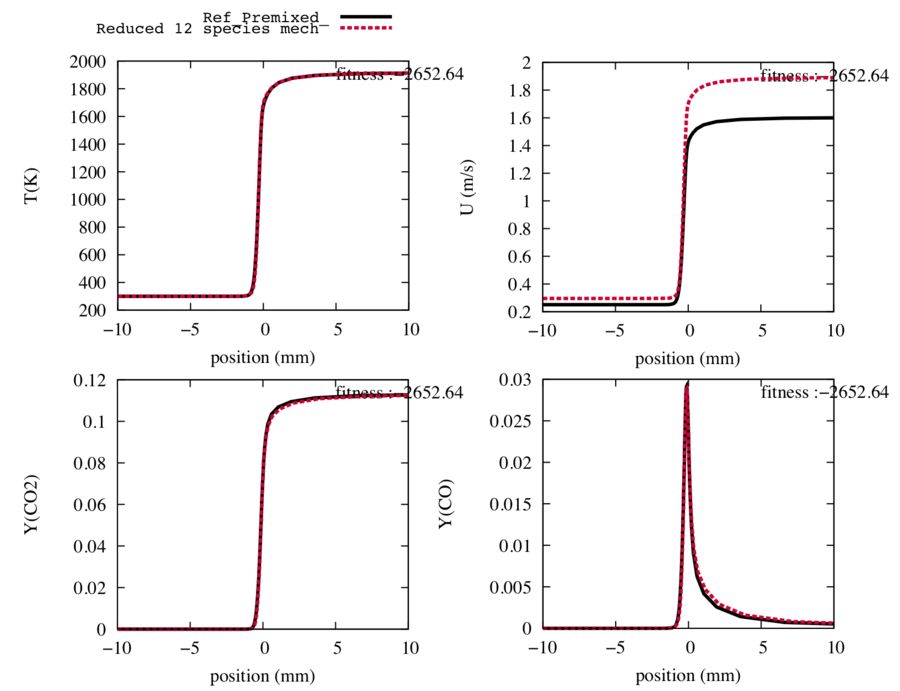
In order to obtain the trajectories of the 14-species 26-reaction reduced scheme with these 2 species in QSS hypothesis, we run the '''getQSSfile''' step and we obtain the following graphs.We observe that the final state and the shape of the trajectories are conserved. It is under these conditions that the optimisation step will be efficient. | In order to obtain the trajectories of the 14-species 26-reaction reduced scheme with these 2 species in QSS hypothesis, we run the '''getQSSfile''' step and we obtain the following graphs.We observe that the final state and the shape of the trajectories are conserved. It is under these conditions that the optimisation step will be efficient. | ||
| + | |||
| + | We can note that the main target profiles are not impacted by the QSS hypothesis, nevertheless the fitness seems better. This is due to errors compensations by the other species not shown here (remind that the fitness is calculated for all species, velocity and temperature, the target just have a larger impact coefficient on the fitness). | ||
[[File:QSS_12sp_Premixed.png|900px|center]] | [[File:QSS_12sp_Premixed.png|900px|center]] | ||
| Line 196: | Line 208: | ||
=== Optimisation === | === Optimisation === | ||
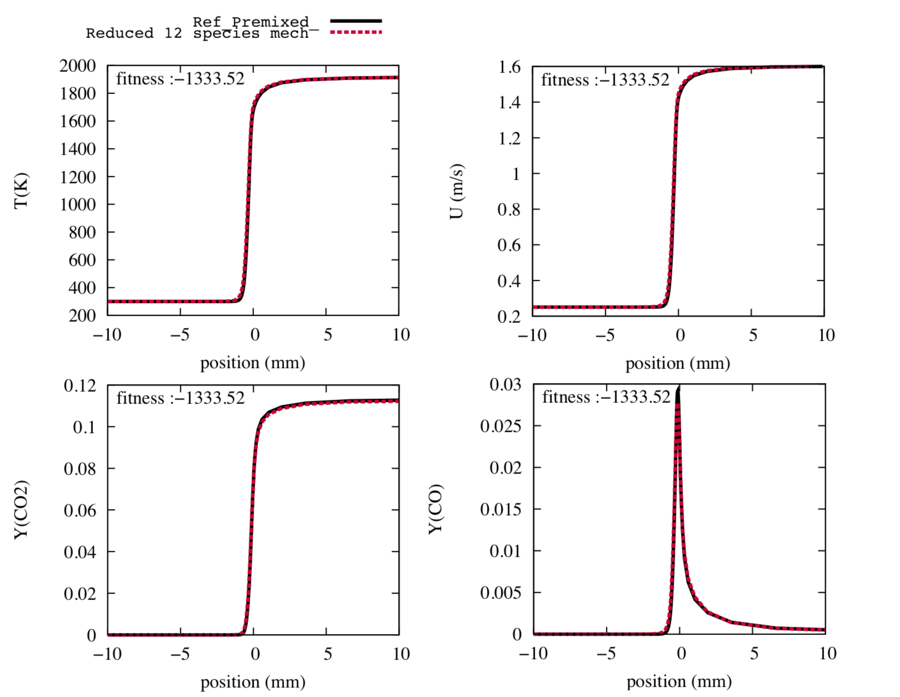
| − | The final step of the ORCh method, the genetic algorithm, enables to recover the trajectories of the target species. Firstly, we allowed a 10 % variation on the pre exponential factor, the temperature exposant and 6 % on the energy activation in order to find their optimal values. | + | The final step of the ORCh method, the genetic algorithm, enables to recover the trajectories of the target species. Firstly, we allowed a 10 % variation on the pre exponential factor, the temperature exposant and 6 % on the energy activation in order to find their optimal values (i.e., <code>AllowedVariation_A = 0.10; AllowedVariation_b = 0.10; AllowedVariation_E = 0.06</code>). |
| − | A population of | + | A population of 40 elements was used during 50 generations. The optimisation step supports running in parallel. For example, we can use 20 processors via <code>mpirun -np 20 mainprogram</code>. Just make sure that PopSize divided by the number of processes is an integer. This step may take a long time, like several hours, depending on how many processors are used. The optimised scheme is in <code>analytic_schemes/Ref/</code> while the corresponding EPS figures can be found in <code>analytic_schemes/PLOTS/</code>. |
| − | |||
The following trajectories of the temperature, the flame speed and the target species match perfectly the ones of the reference detailed scheme with only 12 transported species and 26 reactions. | The following trajectories of the temperature, the flame speed and the target species match perfectly the ones of the reference detailed scheme with only 12 transported species and 26 reactions. | ||
Latest revision as of 16:17, 6 February 2019
Contents
Objectives
The Stochastic_GRI12 test case describes a reduction of the GRI1.2 scheme for a 1D premixed flame. Starting with 32 species and 177 reactions, we reduce to 14 species and 26 reactions.
Key parameters
The target species considered for this test case are O2, CO and CO2. The characteristics of the premixed flame are displayed below (part of the input_file.ini) :
//------Flame parameters------// //Flame 0 Tf = 300 To = 300 Pressure = 1E+05 Equivalence_ratio = 0.75 Xf = CH4:1.0 Xo = O2:0.21, N2:0.79 // the composition can also be added with mass fractions, for that replace "Xf" by "Yf" and "Xo" by "Yo" Initial_flame = flames/flame__Phi_0_75__P_100000__T_300.cantera Final_flame = flames/flame //End
Results
DRGEP step
While running the DRGEP species step (with step = DRGEP_Species in the "input_file.ini"), your terminal should display the following information :
MECHANISM:------------------------------------------------------------------------------------------------------
Reading initial mechanism "mechanisms/gri12.xml" with description "gri12" ----------> OK
Number of species: 32
Number of reactions: 177
PREMIXED FLAME:--------------------------------------------------------------
Reading initial flame "flames/flame__Phi_0_75__P_100000__T_300.cantera" with description "st_flame" ----------> description: test flame
OK
Pressure: 100000
Equivalence ratio: 0.75
Composition (mass fractions):
<O2:0.223145>
<CH4:0.0419532>
<N2:0.734901>
followed by the the species associated with their rank :
-------DRGEP coefficients------- -------------------------------- 1.37642e-15 AR 2.12795e-06 HCCOH 4.32935e-05 C2H 0.000157543 CH2CO 0.000362521 C 0.000444401 C2H2 0.00101888 HCCO 0.00357515 C2H3 0.00398744 CH3OH 0.00409277 CH 0.00543294 CH2OH 0.0126485 C2H4 0.0126485 C2H5 0.0170844 CH2(S) 0.0190984 C2H6 0.0314311 CH2 0.0362943 H2O2 0.0844714 H2 0.0862782 CH3 0.105794 CH2O 0.110349 HCO 0.141159 H2O 0.150828 CH3O 0.218864 O 0.294638 HO2 0.48461 OH 0.48475 H 1 O2 1 CH4 1 CO 1 CO2 1 N2
From a detailed 32 species scheme, we obtain reduced schemes (31 to 14 species). and we choose the one with 14 species for the next step because the shape of the velocity and species are conserved (and Cantera is unable to converge the 13-species scheme. The code will therefore end with an error message. But it is fine). The following Figure 1 is available in the directory outputs/Premixed/ with the file format of EPS.
Fig1 : Comparison between the reference trajectories of the target species, the temperature and the flame speed (in black), and the trajectories computed with the reduced mechanism with 14 transported species and 42 reactions, after a DRGEP species reduction (in red).
From the reduced scheme with 14 species, we then perform the next DRGEP Reactions step by manually change the value of the "step" keyword in the "input_file.ini" as step = DRGEP_Reactions. Make sure the reference mechanism and trajectory are correctly set up (i.e., mech_ref = mechanisms/gri12.xml; trajectory_ref = Ref_DRGEP_Species32). The DRGEP Reactions step displays the associated 42 reactions (forward, reverse and global) with their rank :
Reaction 22 9.23227e-06 Reaction 1 2.08238e-05 Reaction 11 2.29985e-05 Reaction 2 5.30076e-05 Reaction 12 0.00029274 Reaction 18 0.00191785 Reaction 6 0.00198407 Reaction 36 0.00293471 Reaction 38 0.00420356 Reaction 31 0.00462285 Reaction 34 0.00611017 Reaction 7 0.00905302 Reaction 10 0.00921383 Reaction 32 0.0106164 Reaction 37 0.0133867 Reaction 8 0.0136681 Reaction 25 0.0237175 Reaction 24 0.0277978 Reaction 29 0.0305413 Reaction 13 0.0329515 Reaction 9 0.0342601 Reaction 3 0.0398655 Reaction 5 0.0422524 Reaction 14 0.0442348 Reaction 19 0.0493222 Reaction 35 0.102885 Reaction 40 0.103946 Reaction 26 0.111933 Reaction 30 0.131232 Reaction 23 0.159996 Reaction 21 0.161005 Reaction 20 0.180048 Reaction 39 0.18465 Reaction 42 0.19675 Reaction 4 0.208145 Reaction 15 0.215404 Reaction 16 0.222982 Reaction 27 0.274358 Reaction 17 0.274617 Reaction 33 0.403162 Reaction 41 0.437888 Reaction 28 0.438952
After this step, by comparing the reference trajectories with the new ones, we choose to delete 16 reactions because after 17 reactions removed, the CO2 profile loose its initial shape.
The next step is performed with 14 species and 26 reactions.
Fig2 : Comparison between the trajectories of the target species, the temperature and the flame speed computed with the reference 32-species 177-reaction mechanism (in black) , and the trajectories computed with the reduced mechanism with 14 transported species and 26 reactions after a DRGEP reaction reduction (in red).
QSS step
The computeQSSCriteria step provides the QSS criteria of each species and the different links with the others species :
Species H 0.0752034 Species O 0.0211171 Species O2 0.33194 Species OH 0.019287 Species H2O 0.379235 Species HO2 0.0113201 Species CH3 0.107792 Species CH4 0.79541 Species CO 0.693792 Species CO2 0.720209 Species HCO 0.00321461 Species CH2O 0.0817911 Species CH3O 0.0360026 Species N2 0
Interactions with species H with QSS Criteria 0.0752034 HO2:2 CH3:1 CH2O:2 CH3O:1 N2:1
Interactions with species O with QSS Criteria 0.0211171 OH:1 HO2:1 CH3:1 CH2O:1 CH3O:1
Interactions with species OH with QSS Criteria 0.019287 O:1 OH:2 HO2:1 CH3:2 HCO:2 CH2O:1 CH3O:1
Interactions with species HO2 with QSS Criteria 0.0113201 H:2 O:1 OH:1 CH3:1 CH2O:1 N2:1
Interactions with species CH3 with QSS Criteria 0.107792 H:1 O:1 OH:2 HO2:1
Interactions with species HCO with QSS Criteria 0.00321461 OH:2
Interactions with species CH2O with QSS Criteria 0.0817911 H:2 O:1 OH:1 HO2:1
Interactions with species CH3O with QSS Criteria 0.0360026 H:1 O:1 OH:1
Interactions with species N2 with QSS Criteria 0 H:1 HO2:1
We choose to put the species CH3O and HCO in QSS hypothesis due to their low QSS coefficient. The species HO2 could have been a good candidate too but it is linked to himself (non linearity).
In order to obtain the trajectories of the 14-species 26-reaction reduced scheme with these 2 species in QSS hypothesis, we run the getQSSfile step and we obtain the following graphs.We observe that the final state and the shape of the trajectories are conserved. It is under these conditions that the optimisation step will be efficient.
We can note that the main target profiles are not impacted by the QSS hypothesis, nevertheless the fitness seems better. This is due to errors compensations by the other species not shown here (remind that the fitness is calculated for all species, velocity and temperature, the target just have a larger impact coefficient on the fitness).
Fig3 : Comparison between the reference trajectories of the target species, the temperature and the flame speed (in black), and the trajectories computed with the reduced mechanism with 12 transported species and 26 reactions after a DRGEp reaction reduction (in red).
Optimisation
The final step of the ORCh method, the genetic algorithm, enables to recover the trajectories of the target species. Firstly, we allowed a 10 % variation on the pre exponential factor, the temperature exposant and 6 % on the energy activation in order to find their optimal values (i.e., AllowedVariation_A = 0.10; AllowedVariation_b = 0.10; AllowedVariation_E = 0.06).
A population of 40 elements was used during 50 generations. The optimisation step supports running in parallel. For example, we can use 20 processors via mpirun -np 20 mainprogram. Just make sure that PopSize divided by the number of processes is an integer. This step may take a long time, like several hours, depending on how many processors are used. The optimised scheme is in analytic_schemes/Ref/ while the corresponding EPS figures can be found in analytic_schemes/PLOTS/.
The following trajectories of the temperature, the flame speed and the target species match perfectly the ones of the reference detailed scheme with only 12 transported species and 26 reactions.
Fig4 : Comparison between the reference trajectories of the target species, the temperature and the flame speed (in black), and the trajectories computed with the reduced mechanism with 12 transported species and 26 reactions after optimisation (in red).