Difference between revisions of "Multi-inlet CH 4/Air flame with scheme GRI12"
(→Optimisation Step) |
(→DRGEP Step) |
||
| (15 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== '''Objectives''' == | == '''Objectives''' == | ||
| − | The Stochastic_GRI12 test case describes a reduction of the GRI1.2 scheme for a 0D 2-inlet configuration. Starting with 32 species and 177 reactions, we reduce to 12 transported species, 2 species in quasi steady assumption and | + | The Stochastic_GRI12 test case describes a reduction of the GRI1.2 scheme for a 0D 2-inlet configuration. Starting with 32 species and 177 reactions, we reduce to 12 transported species, 2 species in quasi steady state assumption and 28 reactions. |
== '''Key parameters''' == | == '''Key parameters''' == | ||
| − | The target species considered for this test case are O2, CO and CO2. We are at atmospheric pressure, and the fuel is injected in its gaseous form (no evaporation model). The inputs of the 3 inlets (oxidiser, fuel and burned gases) are displayed below : | + | The target species considered for this test case are O2, CO and CO2. We are at atmospheric pressure, and the fuel is injected in its gaseous form (no evaporation model). The inputs of the 3 inlets (oxidiser, fuel and burned gases) are displayed below (from the input_file.ini): |
| − | + | //------Inlets------// | |
| − | + | new Inlet 0 : Air | |
| − | + | T = 700 | |
| − | + | Pressure = 1E+05 | |
| − | + | MassFlowRate = 0.500 | |
| − | + | Xk = O2:0.21, N2:0.79 | |
| − | + | // the composition can also be added with molar fractions, for that replace "Yk" by "Xk" | |
| − | + | EvaporationModel = false | |
| − | + | DropletDiameter = 0.0 | |
| − | + | EvaporationTime = 0.0 | |
| − | + | liquidDensity = 0.0 | |
| − | + | EvaporationLatentHeat = 0.0 | |
| + | //EndInlet | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | new Inlet 1 : Fuel | |
| − | + | T = 450 | |
| − | + | Pressure = 1E+05 | |
| − | + | MassFlowRate = 0.017 | |
| − | + | Xk = CH4:1.0 | |
| − | + | // the composition can also be added with molar fractions, for that replace "Yk" by "Xk" | |
| − | + | EvaporationModel = 0 | |
| − | + | DropletDiameter = 0.0 | |
| − | + | EvaporationTime = 0.0 | |
| − | + | liquidDensity = 0.0 | |
| − | + | EvaporationLatentHeat = 0.0 | |
| − | + | //EndInlet | |
| − | + | ||
| − | + | ||
| − | + | new Inlet : GB | |
| − | + | T = 2500 | |
| + | Pressure = 1E+05 | ||
| + | MassFlowRate = 0.2 | ||
| + | Xk = N2:0.76308, O2:0.093573, H2O:0.072355, CO2:0.070468 | ||
| + | // the composition can also be added with molar fractions, for that replace "Yk" by "Xk" | ||
| + | EvaporationModel = 0 ///1 to activate the model | ||
| + | DropletDiameter = 0.0 | ||
| + | EvaporationTime = 0.0 | ||
| + | liquidDensity = 0.0 | ||
| + | EvaporationLatentHeat = 0.0 | ||
== '''Results''' == | == '''Results''' == | ||
| Line 57: | Line 55: | ||
While running the DRGEP species step, your terminal should display the following information : | While running the DRGEP species step, your terminal should display the following information : | ||
| − | Reading initial mechanism "mechanisms/gri12.xml" with description "gri12" ----------> OK | + | |
| − | + | ||
| − | + | MECHANISM:------------------------------------------------------------------------------------------------------ | |
| − | + | Reading initial mechanism "mechanisms/gri12.xml" with description "gri12" ----------> OK | |
| − | + | Number of species: 32 | |
| − | + | Number of reactions: 177 | |
Composition to enter For the equilibrium computation to get the Burned gases | Composition to enter For the equilibrium computation to get the Burned gases | ||
| − | Compo_H_mixed | + | Compo_H_mixed 525637 |
X_O2: 0.16931 | X_O2: 0.16931 | ||
X_H2O: 0.019848 | X_H2O: 0.019848 | ||
| Line 72: | Line 70: | ||
X_CO2: 0.0193304 | X_CO2: 0.0193304 | ||
X_N2: 0.749689 | X_N2: 0.749689 | ||
| − | T_mixed | + | T_mixed 1212.8 |
Nb particles 0 500 | Nb particles 0 500 | ||
Nb particles 1 17 | Nb particles 1 17 | ||
| Line 85: | Line 83: | ||
0 AR | 0 AR | ||
| − | + | 1.41837e-05 HCCOH | |
| − | + | 0.000287651 C | |
| − | + | 0.000489111 CH2CO | |
| − | + | 0.000498049 C2H | |
| − | + | 0.00327403 C2H2 | |
| − | + | 0.00356528 CH2OH | |
| − | + | 0.00476023 CH | |
| − | + | 0.00528262 HCCO | |
| − | + | 0.0119242 H2O2 | |
| − | 0. | + | 0.0130846 C2H3 |
| − | 0. | + | 0.0164568 CH3OH |
| − | 0. | + | 0.0358106 C2H4 |
| − | 0. | + | 0.0358106 C2H5 |
| − | 0. | + | 0.0380984 CH2(S) |
| − | 0. | + | 0.0394904 CH2 |
| − | 0. | + | 0.0692651 H2 |
| − | 0. | + | 0.120645 C2H6 |
| − | 0. | + | 0.144271 HCO |
| − | 0. | + | 0.146106 CH2O |
| − | 0. | + | 0.193431 CH3O |
| − | 0. | + | 0.345988 O |
| − | 0. | + | 0.699261 H |
| − | + | 0.699262 OH | |
0.999998 HO2 | 0.999998 HO2 | ||
0.999999 CH3 | 0.999999 CH3 | ||
| Line 116: | Line 114: | ||
1 CO2 | 1 CO2 | ||
1 N2 | 1 N2 | ||
| − | |||
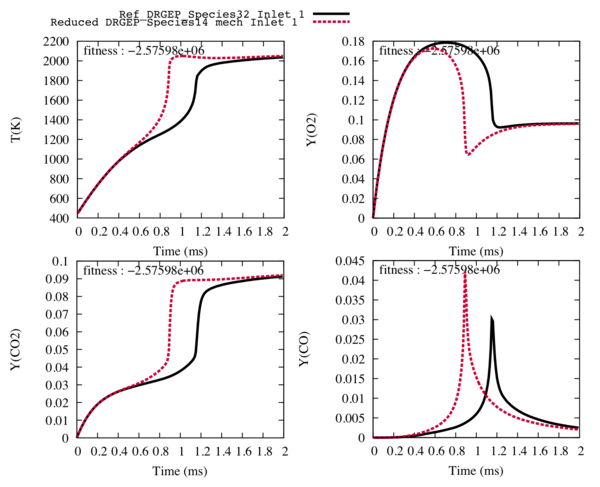
From a detailed 32 species scheme, we obtain reduced schemes (31 to 11 species). and we choose the one with 14 species for the next step. With 13 species and less, the trajectories of the targets and the temperature are too different from the detailed ones to be easily optimised. | From a detailed 32 species scheme, we obtain reduced schemes (31 to 11 species). and we choose the one with 14 species for the next step. With 13 species and less, the trajectories of the targets and the temperature are too different from the detailed ones to be easily optimised. | ||
| Line 126: | Line 123: | ||
From the reduced scheme with 14 species, the DRGEP reaction step displays the associated 42 reactions (forward, reverse and global) with their rank. The global reactions are displayed below : | From the reduced scheme with 14 species, the DRGEP reaction step displays the associated 42 reactions (forward, reverse and global) with their rank. The global reactions are displayed below : | ||
| + | |||
| + | Reaction 11 7.11815e-05 | ||
| + | Reaction 2 9.22955e-05 | ||
| + | Reaction 1 9.5273e-05 | ||
| + | Reaction 18 0.002098 | ||
| + | Reaction 31 0.00268202 | ||
| + | Reaction 10 0.00415073 | ||
| + | Reaction 6 0.00453937 | ||
| + | Reaction 12 0.0058625 | ||
| + | Reaction 22 0.00631152 | ||
| + | Reaction 7 0.0107781 | ||
| + | Reaction 8 0.0107783 | ||
| + | Reaction 34 0.0154049 | ||
| + | Reaction 19 0.0177938 | ||
| + | Reaction 29 0.0220001 | ||
| + | Reaction 25 0.0269477 | ||
| + | Reaction 24 0.0322283 | ||
| + | Reaction 37 0.0341651 | ||
| + | Reaction 40 0.0553785 | ||
| + | Reaction 9 0.0611019 | ||
| + | Reaction 32 0.0701004 | ||
| + | Reaction 39 0.0765675 | ||
| + | Reaction 38 0.148919 | ||
| + | Reaction 35 0.164745 | ||
| + | Reaction 3 0.165682 | ||
| + | Reaction 14 0.166509 | ||
| + | Reaction 30 0.173203 | ||
| + | Reaction 23 0.250228 | ||
| + | Reaction 21 0.364056 | ||
| + | Reaction 36 0.459012 | ||
| + | Reaction 26 0.491301 | ||
| + | Reaction 5 0.494067 | ||
| + | Reaction 28 0.550285 | ||
| + | Reaction 20 0.606395 | ||
| + | Reaction 42 0.607008 | ||
| + | Reaction 15 0.736774 | ||
| + | Reaction 17 0.754281 | ||
| + | Reaction 16 0.791332 | ||
| + | Reaction 41 0.855392 | ||
| + | Reaction 33 0.948061 | ||
| + | Reaction 13 0.969153 | ||
| + | Reaction 4 0.974258 | ||
| + | Reaction 27 0.992507 | ||
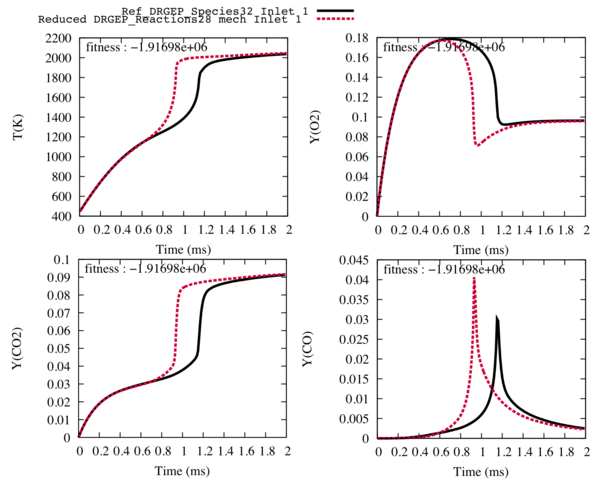
| + | After this step, by comparing the reference trajectories with the new ones, we choose to delete 13 reactions, so the next step is performed with 14 species and 28 reactions. Indeed, with 27 and less species, the final state of the trajectories is not well predicted and difficult to represent even with the optimisation step. | ||
| − | + | [[File:14sp_28R.png|600px|center]] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | [[File: | + | |
| − | Fig2 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 14 transported species and | + | Fig2 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 14 transported species and 28 reactions (in red) for the fuel inlet. |
=== QSS Step === | === QSS Step === | ||
| Line 182: | Line 177: | ||
While running the QSS step, your terminal should display the following information : | While running the QSS step, your terminal should display the following information : | ||
| − | Species H 0. | + | Species H 0.00794323 |
| − | Species O 0. | + | Species O 0.00490131 |
Species O2 1 | Species O2 1 | ||
| − | Species OH 0. | + | Species OH 0.00320187 |
| − | Species H2O 0. | + | Species H2O 0.0582805 |
| − | Species HO2 0. | + | Species HO2 0.012535 |
| − | Species CH3 0. | + | Species CH3 0.0853549 |
| − | Species CH4 0. | + | Species CH4 0.794206 |
Species CO 1 | Species CO 1 | ||
Species CO2 1 | Species CO2 1 | ||
| − | Species HCO 0. | + | Species HCO 0.00362124 |
| − | Species CH2O 0. | + | Species CH2O 0.181152 |
| − | Species CH3O 0. | + | Species CH3O 0.00472456 |
Species N2 1 | Species N2 1 | ||
| − | Interactions with species H with QSS Criteria 0. | + | Interactions with species H with QSS Criteria 0.00794323 |
| − | H2O:2 HO2:1 CH3:1 CH2O:2 CH3O:1 | + | H2O:2 HO2:1 CH3:1 CH2O:2 CH3O:1 |
| − | Interactions with species O with QSS Criteria 0. | + | Interactions with species O with QSS Criteria 0.00490131 |
| − | OH:1 H2O:1 HO2:1 CH3:1 CH2O:1 CH3O:1 | + | OH:1 H2O:1 HO2:1 CH3:1 CH2O:1 CH3O:1 |
| − | Interactions with species OH with QSS Criteria 0. | + | Interactions with species OH with QSS Criteria 0.00320187 |
| − | O:1 OH:2 HO2:1 CH3:2 HCO: | + | O:1 OH:2 HO2:1 CH3:2 HCO:1 CH2O:2 CH3O:1 |
| − | Interactions with species H2O with QSS Criteria 0. | + | Interactions with species H2O with QSS Criteria 0.0582805 |
| − | H:2 O:1 HO2:1 CH3:1 HCO:2 | + | H:2 O:1 HO2:1 CH3:1 HCO:2 |
| − | Interactions with species HO2 with QSS Criteria 0. | + | Interactions with species HO2 with QSS Criteria 0.012535 |
| − | H:1 O:1 OH:1 H2O:1 CH3:2 CH2O:1 | + | H:1 O:1 OH:1 H2O:1 CH3:2 CH2O:1 |
| − | Interactions with species CH3 with QSS Criteria 0. | + | Interactions with species CH3 with QSS Criteria 0.0853549 |
| − | H:1 O:1 OH:2 H2O:1 HO2:2 HCO:1 CH2O:1 | + | H:1 O:1 OH:2 H2O:1 HO2:2 HCO:1 CH2O:1 |
| − | Interactions with species HCO with QSS Criteria 0. | + | Interactions with species HCO with QSS Criteria 0.00362124 |
| − | OH: | + | OH:1 H2O:2 CH3:1 |
| − | Interactions with species CH2O with QSS Criteria 0. | + | Interactions with species CH2O with QSS Criteria 0.181152 |
| − | H:2 O:1 OH:2 HO2:1 CH3:1 | + | H:2 O:1 OH:2 HO2:1 CH3:1 |
| − | Interactions with species CH3O with QSS Criteria 0. | + | Interactions with species CH3O with QSS Criteria 0.00472456 |
H:1 O:1 OH:1 | H:1 O:1 OH:1 | ||
We choose to put the species CH3O and HCO in QSS hypothesis due to their low QSS coefficient. | We choose to put the species CH3O and HCO in QSS hypothesis due to their low QSS coefficient. | ||
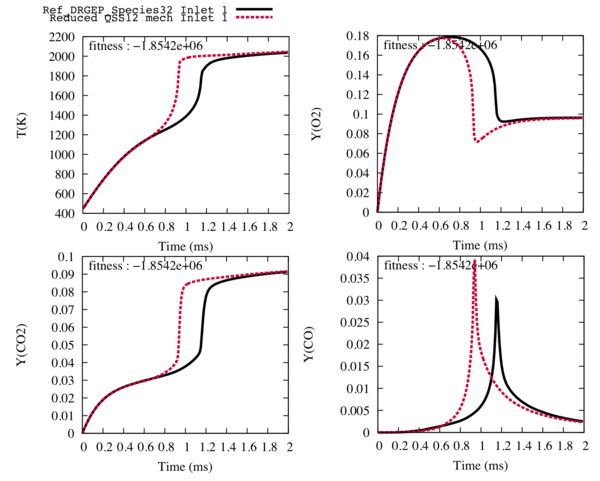
| − | In order to obtain the trajectories of the 12-transported species | + | In order to obtain the trajectories of the 12-transported species 28-reactions reduced scheme, we run the getQSSfile step and we observe that the final state and the shape of the trajectories are conserved. It is under these conditions that the optimisation step will be efficient. |
| − | [[File: | + | [[File:12sp_28R.png|600px|center]] |
| − | Fig3 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 12 transported species and | + | Fig3 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 12 transported species and 28 reactions (in red) for the fuel inlet. |
=== Optimisation Step === | === Optimisation Step === | ||
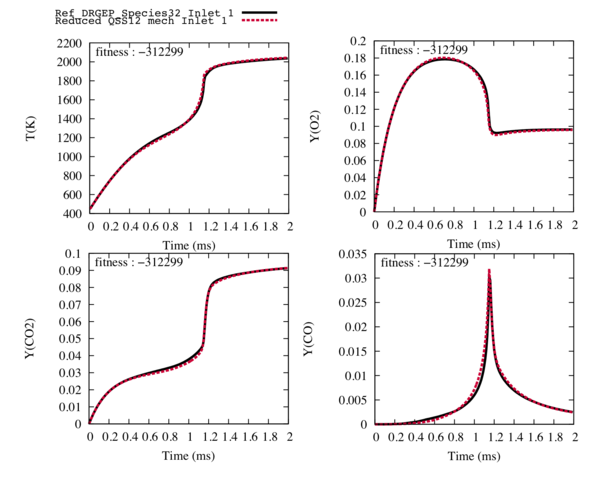
| − | The final step of the ORCh method, the genetic algorithm, enables to recover the trajectories of the target species. | + | The final step of the ORCh method, the genetic algorithm, enables to recover the trajectories of the target species. We allowed a 10 % variation on the pre exponential factor, the temperature exposant and 6 % on the energy activation in order to find their optimal values. |
| − | A population of 40 elements was used during | + | A population of 40 elements was used during 40 generations. |
| − | |||
| − | The following trajectories of the temperature, the flame speed and the target species match perfectly the ones of the reference detailed scheme with only 12 transported species and | + | The following trajectories of the temperature, the flame speed and the target species match perfectly the ones of the reference detailed scheme with only 12 transported species and 28 reactions. |
[[File:optim_stoch_GRI.png|600px|center]] | [[File:optim_stoch_GRI.png|600px|center]] | ||
| − | Fig4 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 12 transported species and | + | Fig4 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 12 transported species and 28 reactions (in red) after optimisation for the fuel inlet. |
Latest revision as of 16:59, 31 January 2019
Objectives
The Stochastic_GRI12 test case describes a reduction of the GRI1.2 scheme for a 0D 2-inlet configuration. Starting with 32 species and 177 reactions, we reduce to 12 transported species, 2 species in quasi steady state assumption and 28 reactions.
Key parameters
The target species considered for this test case are O2, CO and CO2. We are at atmospheric pressure, and the fuel is injected in its gaseous form (no evaporation model). The inputs of the 3 inlets (oxidiser, fuel and burned gases) are displayed below (from the input_file.ini):
//------Inlets------// new Inlet 0 : Air T = 700 Pressure = 1E+05 MassFlowRate = 0.500 Xk = O2:0.21, N2:0.79 // the composition can also be added with molar fractions, for that replace "Yk" by "Xk" EvaporationModel = false DropletDiameter = 0.0 EvaporationTime = 0.0 liquidDensity = 0.0 EvaporationLatentHeat = 0.0 //EndInlet
new Inlet 1 : Fuel T = 450 Pressure = 1E+05 MassFlowRate = 0.017 Xk = CH4:1.0 // the composition can also be added with molar fractions, for that replace "Yk" by "Xk" EvaporationModel = 0 DropletDiameter = 0.0 EvaporationTime = 0.0 liquidDensity = 0.0 EvaporationLatentHeat = 0.0 //EndInlet
new Inlet : GB T = 2500 Pressure = 1E+05 MassFlowRate = 0.2 Xk = N2:0.76308, O2:0.093573, H2O:0.072355, CO2:0.070468 // the composition can also be added with molar fractions, for that replace "Yk" by "Xk" EvaporationModel = 0 ///1 to activate the model DropletDiameter = 0.0 EvaporationTime = 0.0 liquidDensity = 0.0 EvaporationLatentHeat = 0.0
Results
DRGEP Step
While running the DRGEP species step, your terminal should display the following information :
MECHANISM:------------------------------------------------------------------------------------------------------
Reading initial mechanism "mechanisms/gri12.xml" with description "gri12" ----------> OK
Number of species: 32
Number of reactions: 177
Composition to enter For the equilibrium computation to get the Burned gases Compo_H_mixed 525637 X_O2: 0.16931 X_H2O: 0.019848 X_CH4: 0.0418227 X_CO2: 0.0193304 X_N2: 0.749689 T_mixed 1212.8 Nb particles 0 500 Nb particles 1 17 Nb particles 2 200 Nmix 35 Set the mole fraction of inlet 0 Set the mole fraction of inlet 1 Set the mole fraction of inlet 2
followed by the the species associated with their rank :
0 AR 1.41837e-05 HCCOH 0.000287651 C 0.000489111 CH2CO 0.000498049 C2H 0.00327403 C2H2 0.00356528 CH2OH 0.00476023 CH 0.00528262 HCCO 0.0119242 H2O2 0.0130846 C2H3 0.0164568 CH3OH 0.0358106 C2H4 0.0358106 C2H5 0.0380984 CH2(S) 0.0394904 CH2 0.0692651 H2 0.120645 C2H6 0.144271 HCO 0.146106 CH2O 0.193431 CH3O 0.345988 O 0.699261 H 0.699262 OH 0.999998 HO2 0.999999 CH3 1 O2 1 H2O 1 CH4 1 CO 1 CO2 1 N2
From a detailed 32 species scheme, we obtain reduced schemes (31 to 11 species). and we choose the one with 14 species for the next step. With 13 species and less, the trajectories of the targets and the temperature are too different from the detailed ones to be easily optimised.
Fig1 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 14 transported species and 42 reactions (in red) for the fuel inlet.
From the reduced scheme with 14 species, the DRGEP reaction step displays the associated 42 reactions (forward, reverse and global) with their rank. The global reactions are displayed below :
Reaction 11 7.11815e-05 Reaction 2 9.22955e-05 Reaction 1 9.5273e-05 Reaction 18 0.002098 Reaction 31 0.00268202 Reaction 10 0.00415073 Reaction 6 0.00453937 Reaction 12 0.0058625 Reaction 22 0.00631152 Reaction 7 0.0107781 Reaction 8 0.0107783 Reaction 34 0.0154049 Reaction 19 0.0177938 Reaction 29 0.0220001 Reaction 25 0.0269477 Reaction 24 0.0322283 Reaction 37 0.0341651 Reaction 40 0.0553785 Reaction 9 0.0611019 Reaction 32 0.0701004 Reaction 39 0.0765675 Reaction 38 0.148919 Reaction 35 0.164745 Reaction 3 0.165682 Reaction 14 0.166509 Reaction 30 0.173203 Reaction 23 0.250228 Reaction 21 0.364056 Reaction 36 0.459012 Reaction 26 0.491301 Reaction 5 0.494067 Reaction 28 0.550285 Reaction 20 0.606395 Reaction 42 0.607008 Reaction 15 0.736774 Reaction 17 0.754281 Reaction 16 0.791332 Reaction 41 0.855392 Reaction 33 0.948061 Reaction 13 0.969153 Reaction 4 0.974258 Reaction 27 0.992507
After this step, by comparing the reference trajectories with the new ones, we choose to delete 13 reactions, so the next step is performed with 14 species and 28 reactions. Indeed, with 27 and less species, the final state of the trajectories is not well predicted and difficult to represent even with the optimisation step.
Fig2 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 14 transported species and 28 reactions (in red) for the fuel inlet.
QSS Step
While running the QSS step, your terminal should display the following information :
Species H 0.00794323 Species O 0.00490131 Species O2 1 Species OH 0.00320187 Species H2O 0.0582805 Species HO2 0.012535 Species CH3 0.0853549 Species CH4 0.794206 Species CO 1 Species CO2 1 Species HCO 0.00362124 Species CH2O 0.181152 Species CH3O 0.00472456 Species N2 1
Interactions with species H with QSS Criteria 0.00794323 H2O:2 HO2:1 CH3:1 CH2O:2 CH3O:1
Interactions with species O with QSS Criteria 0.00490131 OH:1 H2O:1 HO2:1 CH3:1 CH2O:1 CH3O:1
Interactions with species OH with QSS Criteria 0.00320187 O:1 OH:2 HO2:1 CH3:2 HCO:1 CH2O:2 CH3O:1
Interactions with species H2O with QSS Criteria 0.0582805 H:2 O:1 HO2:1 CH3:1 HCO:2
Interactions with species HO2 with QSS Criteria 0.012535 H:1 O:1 OH:1 H2O:1 CH3:2 CH2O:1
Interactions with species CH3 with QSS Criteria 0.0853549 H:1 O:1 OH:2 H2O:1 HO2:2 HCO:1 CH2O:1
Interactions with species HCO with QSS Criteria 0.00362124 OH:1 H2O:2 CH3:1
Interactions with species CH2O with QSS Criteria 0.181152 H:2 O:1 OH:2 HO2:1 CH3:1
Interactions with species CH3O with QSS Criteria 0.00472456 H:1 O:1 OH:1
We choose to put the species CH3O and HCO in QSS hypothesis due to their low QSS coefficient.
In order to obtain the trajectories of the 12-transported species 28-reactions reduced scheme, we run the getQSSfile step and we observe that the final state and the shape of the trajectories are conserved. It is under these conditions that the optimisation step will be efficient.
Fig3 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 12 transported species and 28 reactions (in red) for the fuel inlet.
Optimisation Step
The final step of the ORCh method, the genetic algorithm, enables to recover the trajectories of the target species. We allowed a 10 % variation on the pre exponential factor, the temperature exposant and 6 % on the energy activation in order to find their optimal values.
A population of 40 elements was used during 40 generations.
The following trajectories of the temperature, the flame speed and the target species match perfectly the ones of the reference detailed scheme with only 12 transported species and 28 reactions.
Fig4 : Comparison between the reference trajectories of the target species and the temperature (in black), and the trajectories computed with the reduced mechanism with 12 transported species and 28 reactions (in red) after optimisation for the fuel inlet.